|
|
|
|
|
|
|
|
|
|
|
Overview
|
|
|
MicroRNAs (miRNAs) are small (19-22 nucleotide) non-protein-coding RNA molecules that function to regulate the expression of specific gene products via hybridization to mRNA transcripts, in concert with associated ribonucleoprotein complexes, to effect translational blockade or message degradation. Although miRNAs have been implicated in numerous developmental and adult disease states, including cancer, their impact on distinct biological pathways and phenotypes remains largely unknown. To address this need for the biomedical research community, we have developed MMIAs, microRNA and mRNA integrated analysis, a versatile and user-friendly web server. MMIA integrates microRNA and mRNA expression data with predicted microRNA target information for analyzing microRNA-associated phenotypes and biological functions by Gene Set Enrichment Analysis (GSEA). To assign biological relevance to the integrated microRNA/mRNA profiles, MMIA uses exhaustive human genome coverage (5782 gene sets), including various disease-associated genes as well as conventional canonical pathways and Gene Ontology. This novel web server provides users with miRNA-mRNA expression data combined analysis tools and broad gene sets.
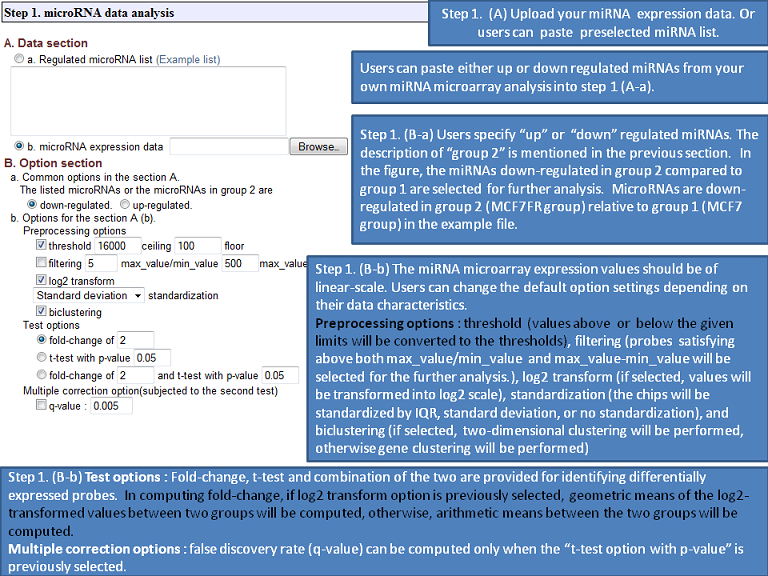
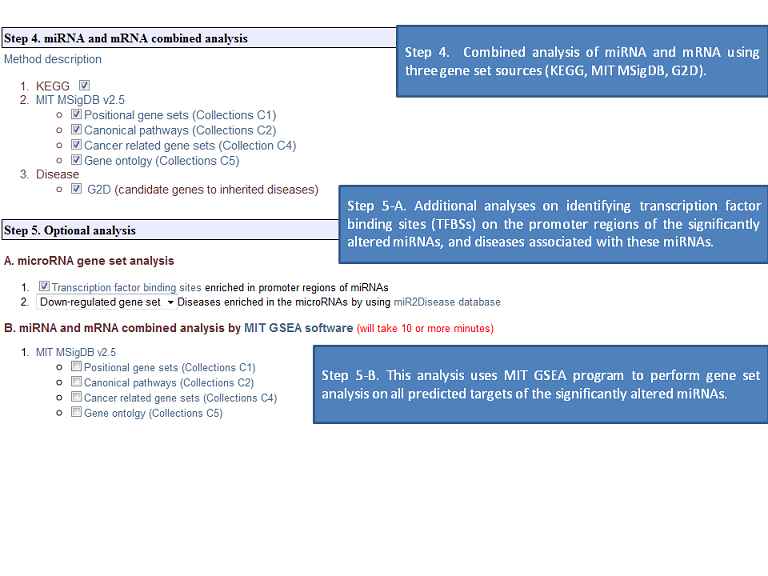
 The expression microRNA and mRNA data input for MMIA is a simple tab-delimited data format, followed by two header lines for class and sample descriptions. To provide further versatility, the user can also enter a regulated microRNA list in the microRNA input field. Expression microarray annotations supported by the server include Ensemble (www.ensembl.org), the NCBI RefSeq and Entrez Gene (www.ncbi.nlm.nih.gov), Swiss-Prot (www.ebi.ac.uk/swissprot), and Affymetrix probesets (www.affymetrix.com). The output shows not only significantly differentially expressed microRNAs and their predicted mRNA target information but also miRNA-mRNA expression combined pathway analysis based on GSEA, as performed for the predefined gene sets in terms of statistically differentially expressed mRNA predicted targets by multiple up/down regulated microRNA genes. Our major gene set is derived from the three universally used sources: 1) KEGG (www.genome.jp/kegg/), 2) MIT MSigDB (www.broad.mit.edu/gsea/msigdb/index.jsp), 3) G2D (www.ogic.ca/projects/g2d_2/), providing information such as biological pathway, cancer-related genes, Gene Ontology, and inherited diseases. Additional novel features of the web server include microRNA gene promoter (in annotated pri-miRNA regions) analysis for conserved transcription factor binding sites (genome.ucsc.edu) and disease-related microRNA database, miR2Disease (www.mir2disease.org). In summary, we have implemented a novel, user-friendly biological analysis web server for extensive biological analysis of microRNA functions. We believe MMIA will be a highly valuable tool for the life science research community.
The expression microRNA and mRNA data input for MMIA is a simple tab-delimited data format, followed by two header lines for class and sample descriptions. To provide further versatility, the user can also enter a regulated microRNA list in the microRNA input field. Expression microarray annotations supported by the server include Ensemble (www.ensembl.org), the NCBI RefSeq and Entrez Gene (www.ncbi.nlm.nih.gov), Swiss-Prot (www.ebi.ac.uk/swissprot), and Affymetrix probesets (www.affymetrix.com). The output shows not only significantly differentially expressed microRNAs and their predicted mRNA target information but also miRNA-mRNA expression combined pathway analysis based on GSEA, as performed for the predefined gene sets in terms of statistically differentially expressed mRNA predicted targets by multiple up/down regulated microRNA genes. Our major gene set is derived from the three universally used sources: 1) KEGG (www.genome.jp/kegg/), 2) MIT MSigDB (www.broad.mit.edu/gsea/msigdb/index.jsp), 3) G2D (www.ogic.ca/projects/g2d_2/), providing information such as biological pathway, cancer-related genes, Gene Ontology, and inherited diseases. Additional novel features of the web server include microRNA gene promoter (in annotated pri-miRNA regions) analysis for conserved transcription factor binding sites (genome.ucsc.edu) and disease-related microRNA database, miR2Disease (www.mir2disease.org). In summary, we have implemented a novel, user-friendly biological analysis web server for extensive biological analysis of microRNA functions. We believe MMIA will be a highly valuable tool for the life science research community.
|
|
|
|
Method
|
|
|
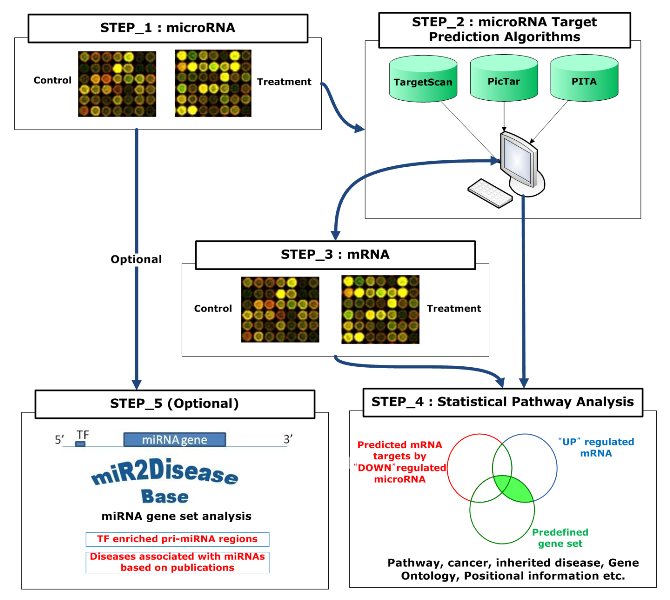
The simple overview of the work flow
 Xin, F., Li, M., Balch, C., Thomson, M., Fan, M., Liu, Y., Hammond, S.M., Kim, S., Nephew, K.P., Computational Analysis of MicroRNA Profiles and Their Target Genes Suggests Significant Involvement in Breast Cancer Antiestrogen resistance, Bioinformatics, 2009, 25:430-434.
Xin, F., Li, M., Balch, C., Thomson, M., Fan, M., Liu, Y., Hammond, S.M., Kim, S., Nephew, K.P., Computational Analysis of MicroRNA Profiles and Their Target Genes Suggests Significant Involvement in Breast Cancer Antiestrogen resistance, Bioinformatics, 2009, 25:430-434.
|
|
|
|
Data format
|
|
|
- General SIP format description
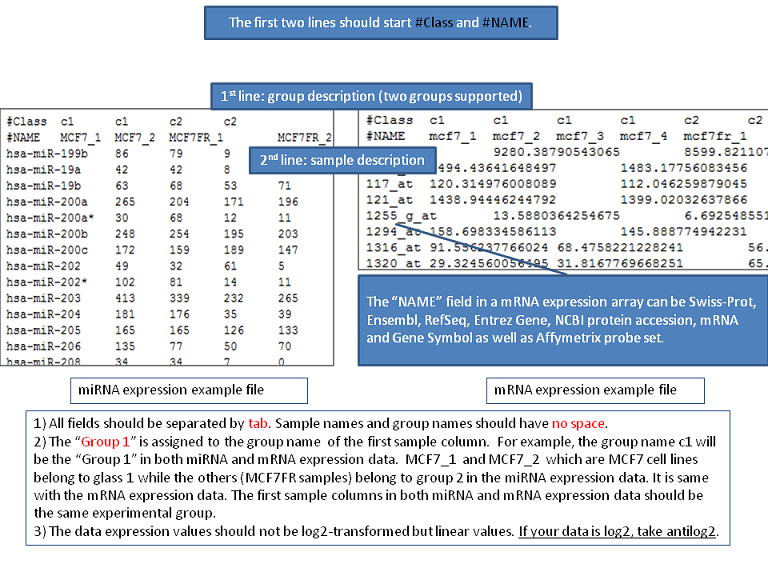
- How to convert log fold-change data in your EXCEL sheet into the SIP format by using Microsoft EXCEL
If fold-change data is used,
i) users leave the the thresholding checkbox ,the filtering checkbox, and the log2 transformation checkbox in the "Preprocessing options" unchecked in miRNA or mRNA data section,
ii) users select "None" in the "standardization" method in miRNA or mRNA data section, and
iii) users check the "fold-change" checkbox in the "Test options" in miRNA or mRNA data section.
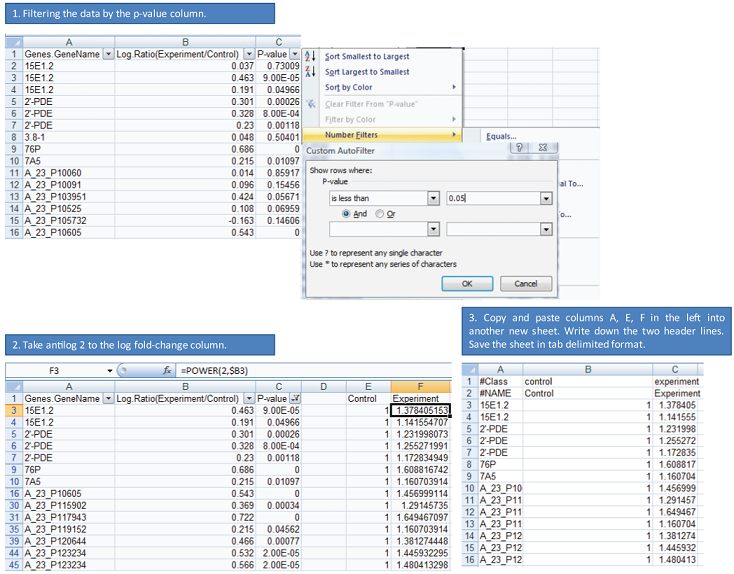
- How to feed pre-selected miRNA list and mRNA list into MMIA
It can be done by making a fake expression data. See the example and its materials (Genes5AzUnstimDwn.sip, Genes5AzUnstimDwn.txt, Genes5AzUnstimDwn.xls, miR_UnstimAll5AzvsNo.txt).
|
|
|
|
Input
|
|
|
|
The main input steps of the server are Steps #1.A Data section and #3.A Data section. The sections need microRNA expression data and mRNA expression data which comply with the format in the previous description. The following is a general procedure.
|
|
|
|
Output (Output samples, click it)
|
|
|
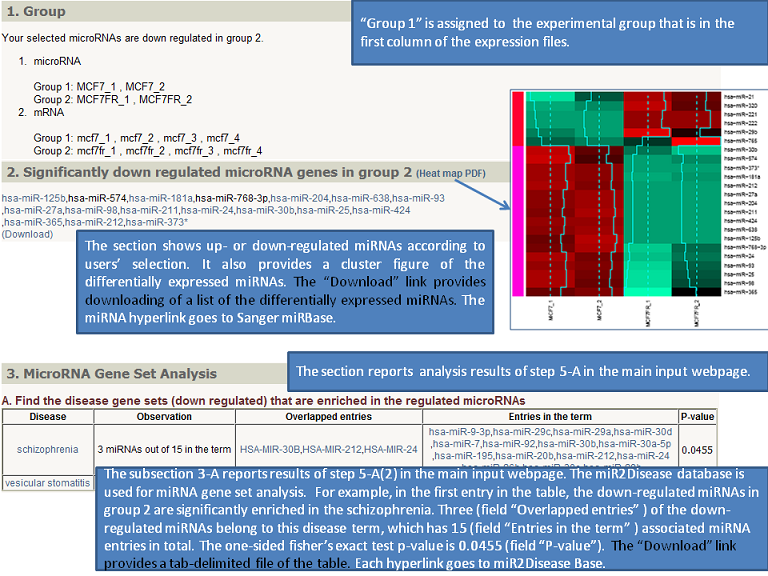
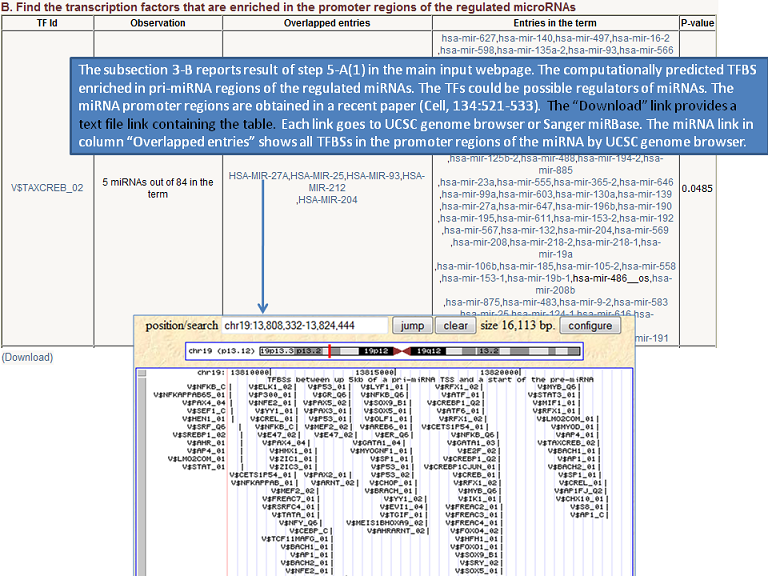
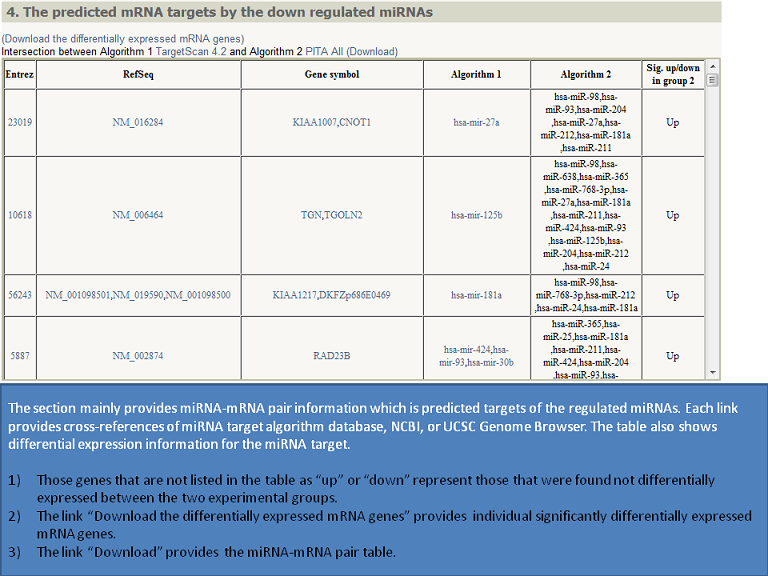
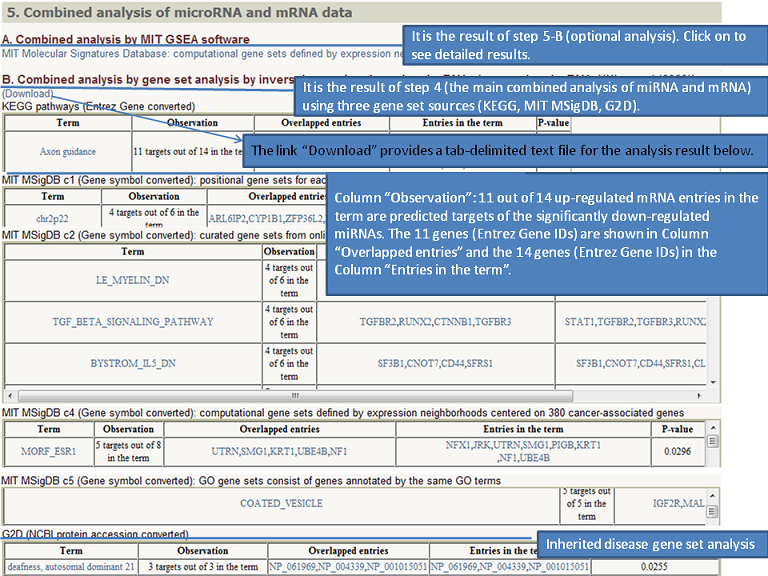
|
|
|
|
|
|
Citation
|
|
|
|
Seungyoon Nam, Meng Li, Kwangmin Choi, Curtis Balch, Sun Kim, and Kenneth P. Nephew, "MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression", Nucleic Acids Research, Web Server Issue (2009) Vol 37, W356-W362.
|
|
|
|
FAQ
|
|
|
|
FAQ
|
|
|
|
Contact
|
|
|
For any bug reports, suggestions and helps, please contact at Dr. Nephew.

|
|
|