Prior Data List
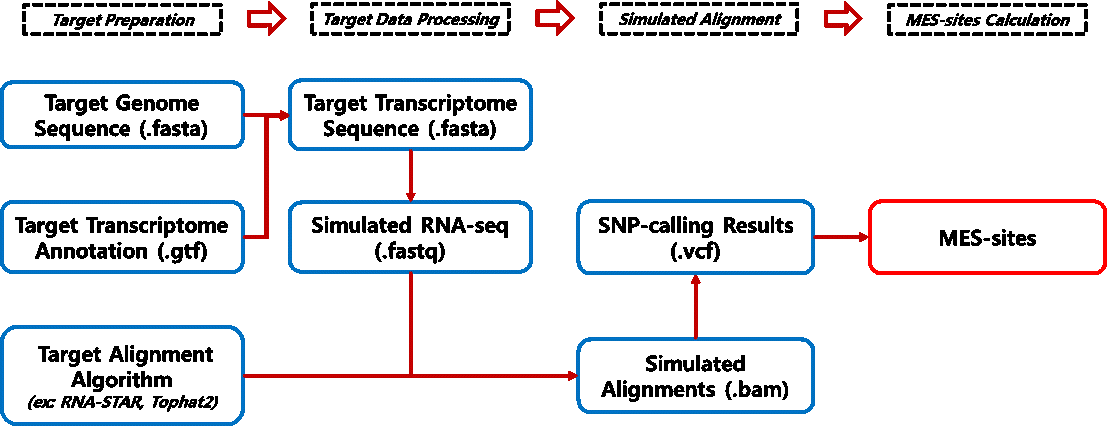
I. Introduction
1) MES method is a simulation-based approach for calculating mapping error-prone locus within genomic sequences (Peng, Zhiyu, et al., 2012).
2) RDDpred utilzed pre-calculated MES-sites of each genomes as prior data.
3) Here we provide pre-calculated results for some model organisms, such as, human, mouse, drosophila, and arabidopsis (RDDpred_MES_Summary.pdf).
II. Human (Homo sapiens)
**Pipeline Overvie & Results Summary: RDDpred_MES_Summary.pdf
1) We selected hg19 genome, NCBI refseq annotation, and RNA-STAR as targets.
2) From given targets, we calculated 16.5-M MES-sites from 10-M simulated reads (Human.MES.txt.gz).
III. Mouse (Mus musculus)
**Pipeline Overvie & Results Summary: RDDpred_MES_Summary.pdf
1) We selected mm10 genome, NCBI refseq annotation, and RNA-STAR as targets.
2) From given targets, we calculated 15-M MES-sites from 10-M simulated reads (Mouse.MES.txt.gz).
IV. Drosophila (Drosophila melanogaster)
**Pipeline Overvie & Results Summary: RDDpred_MES_Summary.pdf
1) We selected dm3 genome, NCBI refseq annotation, and RNA-STAR as targets.
2) From given targets, we calculated 8.3-M MES-sites from 10-M simulated reads (Drosophila.MES.txt.gz).
V. Arabidopsis (Arabidopsis thaliana)
**Pipeline Overvie & Results Summary: RDDpred_MES_Summary.pdf
1) We selected TAIR9 genome, TAIR9 annotation, and RNA-STAR as targets.
2) From given targets, we calculated 11.3-M MES-sites from 10-M simulated reads (Arabidopsis.MES.txt.gz).