TimesVector: A vectorized clustering approach to the analysis of time series transcriptome data from multiple phenotypes
TimesVector is a triclustering tool for clustering time-series data that comprises multiple conditions, or phenotypes. It identifies gene clusters that exhibit distinctly similar or different gene expression patterns among the comparing sample conditions. For example, for a time-series data set of five different strains of yeast, each data sampled at 3 time points, TimesVector will search for gene clusters where a specific strain shows a distinctively different gene expression pattern from the others.
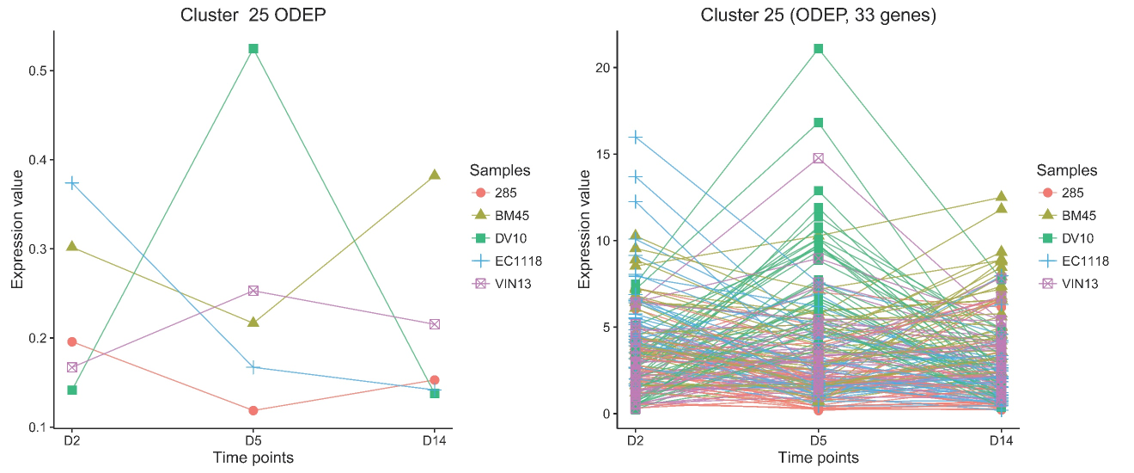
As an example result, 33 genes in cluster 25 were significantly induced at day 5 (D5) during fermentation, only in the DV10 yeast strain (data from GSE11651).
TimesVector is desgined to analyze time-series data of both microarray and NGS gene expression data types. Currently, TimesVector is available only for Linux platforms.
News
- Dec. 23, 2019
- Aug. 03, 2016 Released TimesVector v1.0
Updated TimesVector v1.5: New plots with normalized expression value.