Phase 1 : Prerequisites & Input
- The only prequisite for BioVLAB-MMIA-NGS is JRE.
(1) Click to link JRE install page.
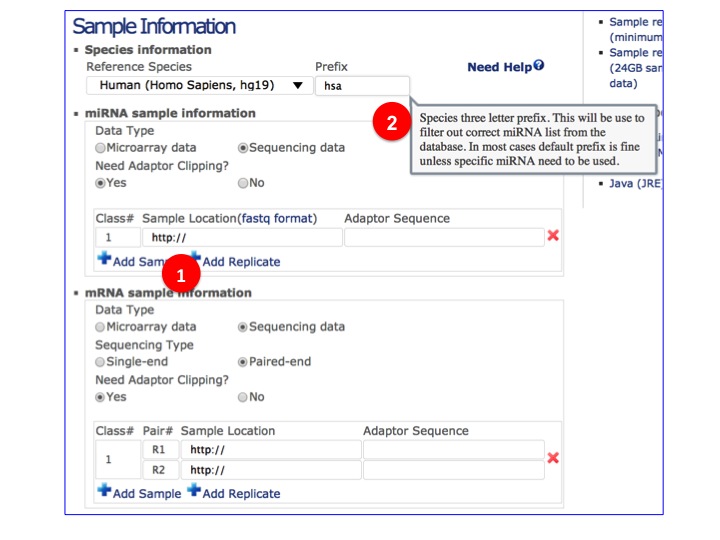
- Select & put sample detail informations.
(1) Click to add sample or replicate.
(2) Move mouse over each field to see the help popup.
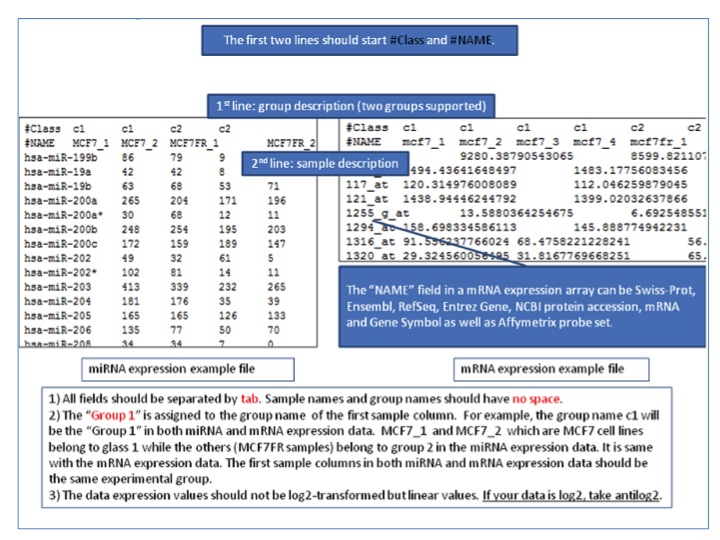
- SIP data format for microarray data
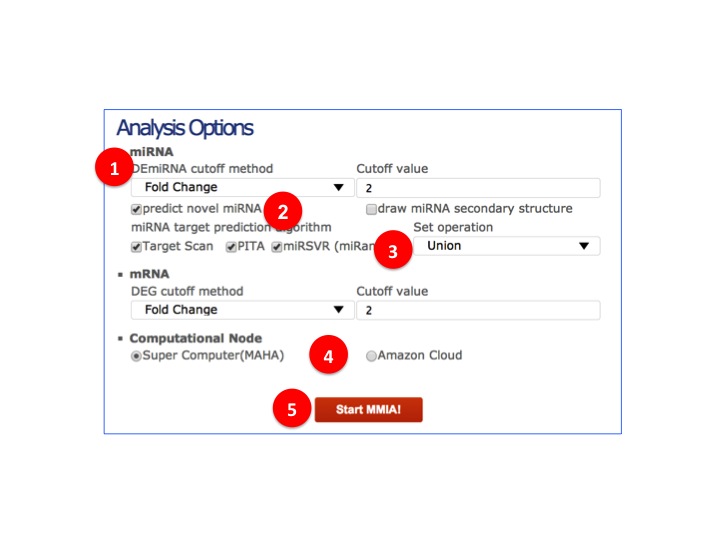
- Detail analysis options.
(1) Method of filtering differential/Significantly expressed miRNAs and cutoff value.
(2) Predicting novel miRNA and drawing secondary structure of miRNAs. Checking both will extend runtime.
(3) MicroRNA target prediction algorithms/databases and way to merge their results.
(4) Computational node to run analysis.
(5) Click to validate inputs & start analysis.
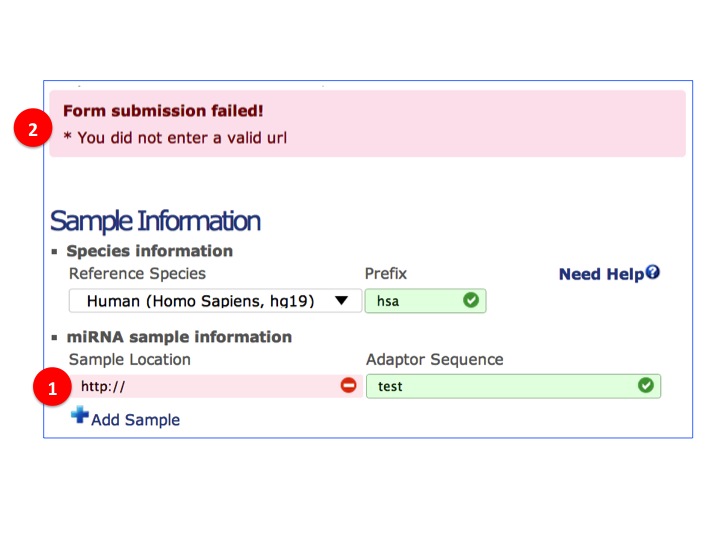
- Field validation
(1) For sample location, user need to input URL. System will retrieve the sample data with it.
(2) Missing/Incorrect data will be notified.
Phase 2 : Invoke XBaya
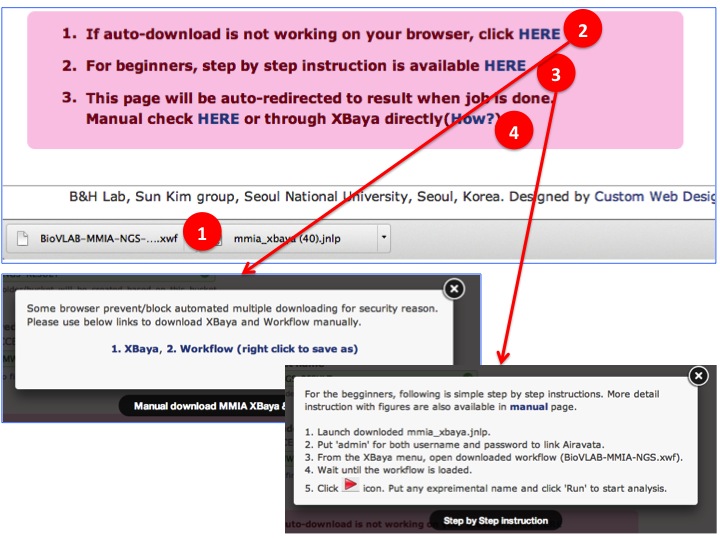
- Once all inputs are validated, constructed workflow based on user inputs are available for downloading.
(1) Auto downloding process(downloading varies depends on browser).
(2) Click to manually download in case automatic downloading functionality is not working on your browser
(3) Invoke XBaya. Step by Step instruction is available for beginners.
(4) Result page link for bookmark is available.
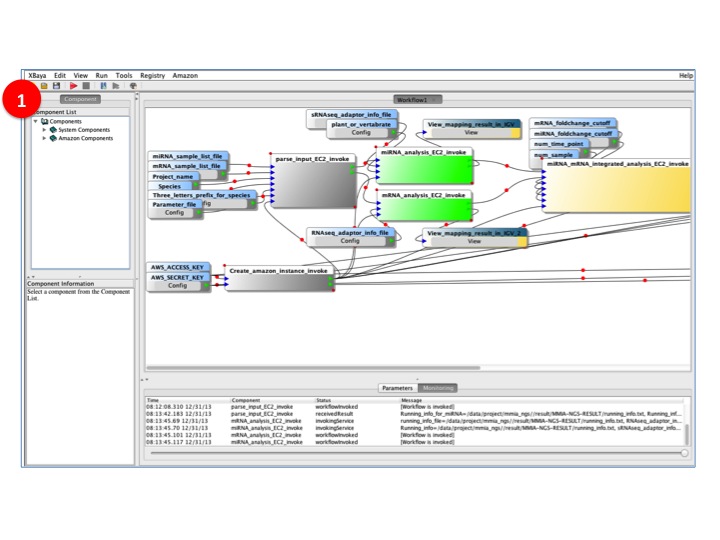
- BioVLAB-MMIA-NGS workflow on the XBaya is running
(1) XBaya main panel shows the progress of the workflow. Gray represents for 'finished' status, Green indicates for currently 'running' status, and Yellow stands for 'ready' status
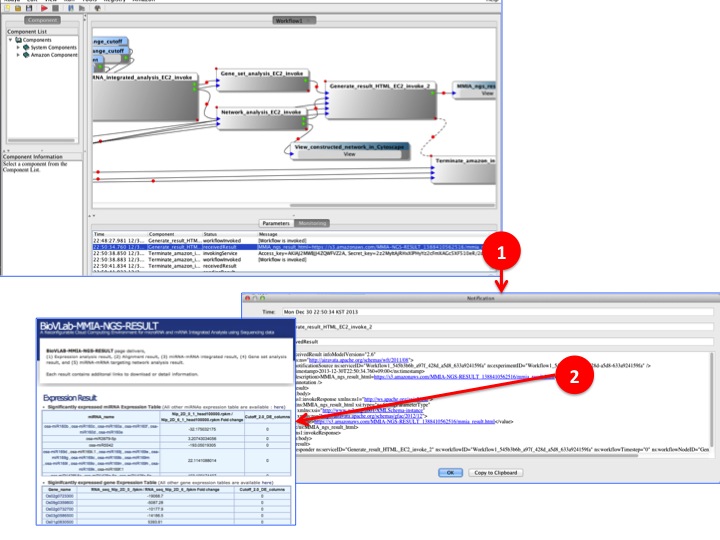
- BioVLAB-MMIA-NGS workflow process is done & Check the result directly from XBaya
(1) Once the process is done, you can find the 'MMIA_ngs_html=XXX" in the monitoring panel. Double click will open notification panel for details.
(2) In the notification panel, there is hyper linked result notification named 'http://epigenomics.snu.ac.kr/..." or 'http://s3.amazonaws.com/...". Click the link to view analysis result.
Phase 3 : Result
- BioVLAB-MMIA-NGS result page contains miRNA analysis result, mRNA analysis result, Integrated analysis result, Gene set analysis result, Network analysis result, and novel miRNA prediction result.
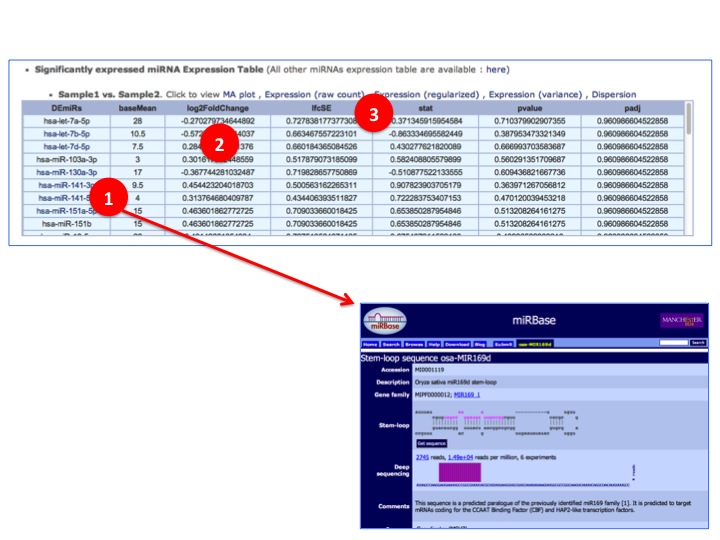
(1) Differentially/Significantly expressed miRNAs (DEmiRs) are listed. Each DEmiR is linked to external database for detail information.
(2) Log2 Fold change between samples which shows expression difference between them.
(3) lftSE: the standard error estimate for the log2 fold change estimate, stat: statistic result, pvalue: t-test pvalue, padj: 'p.adjust' with 'method="BH"' (Benjamini-Hochberg) on the 'pval'.
- Alignment Visualization
(1) Clicking the link will invoke JNLP-based Integrative Genome Browser (IGV) without manual user installation. The alignment result data will be automatically loaded and visualized.
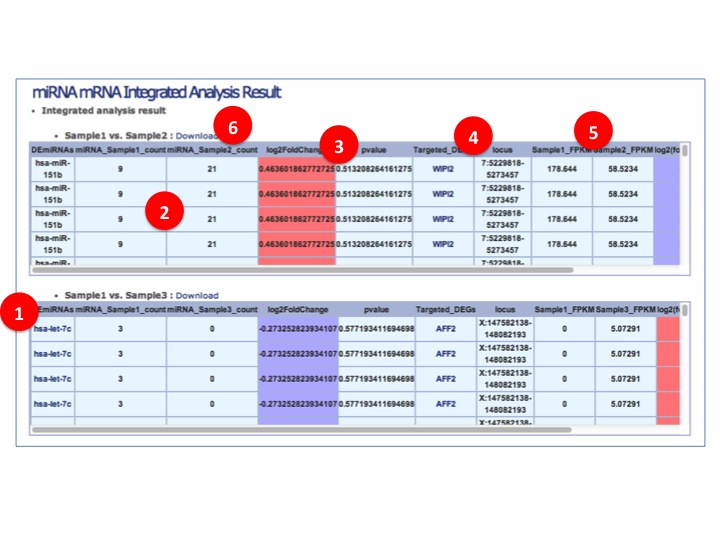
- miRNA-mRNA Integrated Analysis
(1) Differentlly/Significantly expressed miRNAs (DEmiRNAs) are listed.
(2) Samples' expression level (read count)
(3) miRNA fold changes between samples. Up regulated miRNA are colored as red and Down regulated miRNA are colored as blue.
(4) Targeted DEGs and their locus.
(5) Targeted DEGs' expression level
(6) Download whole table
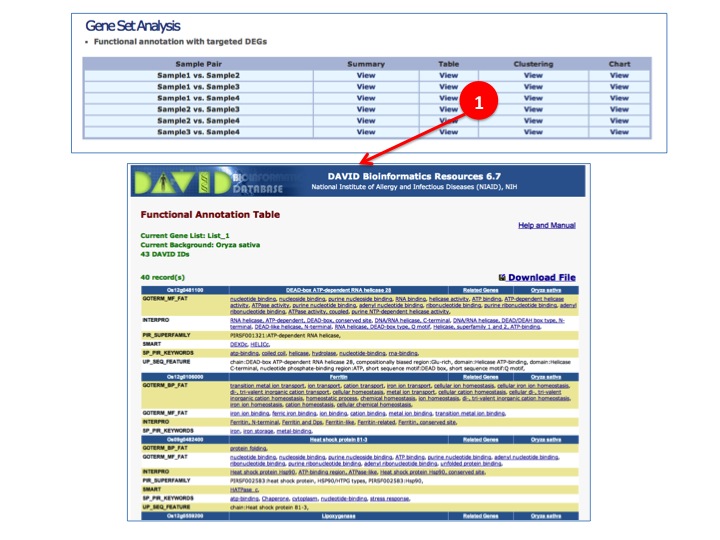
- Gene Set Analysis
(1) DEGs are directly sent to DAVAID for Functional annotation summary, table, clustering, and chart.
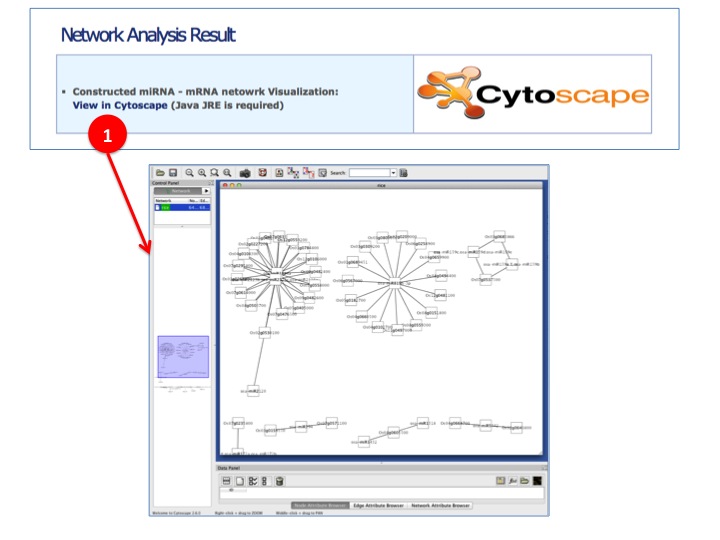
- Network Analysis
(1) Constructed miRNA - mRNA network result is visualized by Cytoscape. Clicking link will invoke JNLP-based Cytoscape without manual user installation. Constructed network is automatically visualized through Cytoscape for additinal miRNA - mRNA targeting network research.
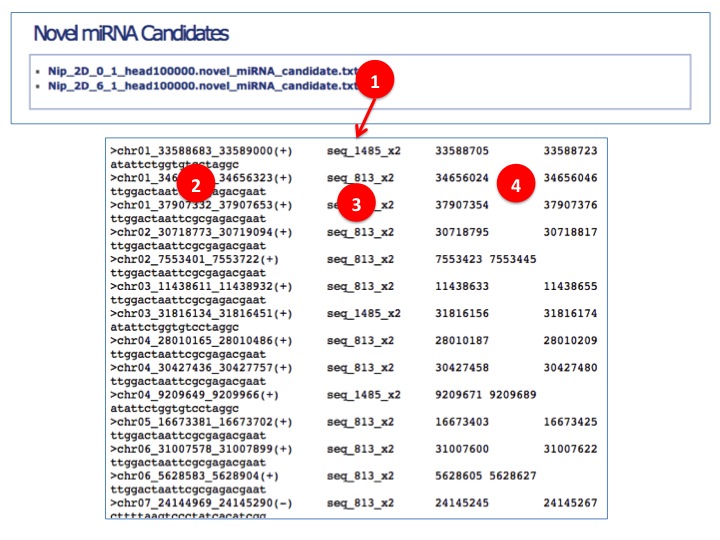
- Novel miRNA Prediction
(1) Click to see the novel miRNA candidates per sample.
(2) Genomic coordinats of predicted primary-miRNA candidates, chromosome_start_end(strand).
(3) Arbitrary read ID with reads count. All the same reads are merged.
(4) Genomic coordinates of predicted miRNA candidates.
Workflow, Method, Pipeline and Running time
- Method
BioVLAB-MMIA-NGS workflow is consist of multiple steps and each step contains several analysis pipelines. We keep the overall method of miRNA-mRNA integrated analysis used in our previous work "Xin, F., Li, M., Balch, C., Thomson, M., Fan, M., Liu, Y., Hammond, S.M., Kim, S., Nephew, K.P., Computational Analysis of MicroRNA Profiles and Their Target Genes Suggests Significant Involvement in Breast Cancer Antiestrogen resistance, Bioinformatics, 2009, 25:430-434.", but for sequencing data analysis, we added various piplines at each steps. Workflow and pipeline details are explained belows.
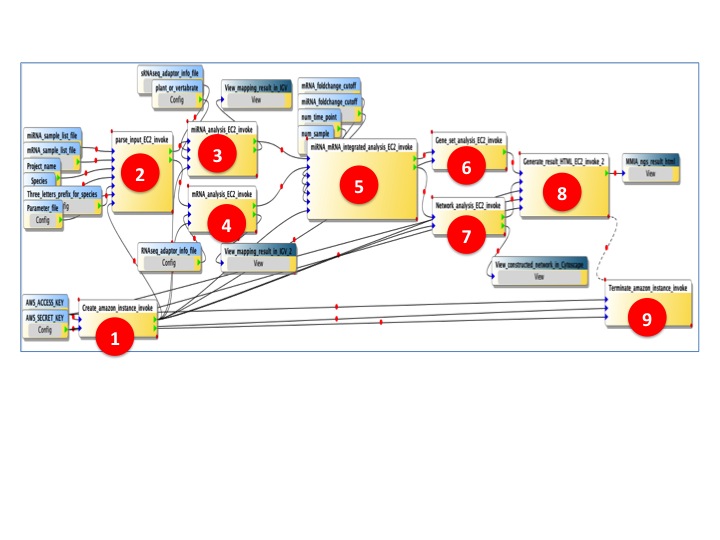
- BioVLAB-MMIA-NGS Workflow
(1) Amazon EC2 instance creation.
(2) Parsing the user inputs, trasfering user data to EC2 instance created in previous step.
(3) microRNA analysis pipeline. To identify miRNAs in smallRNAseq data, we use Bowtie to align reads to miRBase database. For miRNA targeting, we applied sequence matching method between microRNA and mRNA which is used in miRDeep2 for vertabrate and miREvo for plant. For microarray data, we applied same method used in our previous work MMIA.
(4) mRNA analysis pipeline. To analyze RNAseq data, we apply Tophat-Cufflinks pipeline to find DEGs.
(5) microRNA - mRNA integrated analysis. Differentially expressed miRNAs (DEmiR) and targeted DEGs are integrated.
(6) Gene Set Analysis (GSA). Based on DEGs targeted by DEmiRs, automatically construct DAVID query to retrive result of GSA.
(7) microRNA - mRNA targeting network analysis. Based on the targeting information between microRNAs and mRNAs, construct network topology and visualize by Cytoscape.
(8) Automatically generate result page which contains all result of workflow.
(9) After all the workflow is procssed, EC2 instance is terminated.
|
- BioVLAB-MMIA-NGS Running time
In BioVLAB-MMIA-NGS workflow, the pipelines between and within stpes are parallized by utilizing multicore processes. In MAHA, a Super computing environment, jobs are running uder SLURM. Each node has 16cores with 64GB memory. BioVLAB-MMIA-NGS analysis with 50GB (sample data set used on the website), the whole analysis takes 9hours to be completed except data trasfering time which quite varies depends on their location. In Amazon Cloud, with C1.xlarge EC2 instance (vCPU:8, Memory 7GB), and NGS dataset total 24GB {(RNAseq, 5GB each, paired-end, 2 samples : total 20GB), (smallRNAseq, 2GB each, single-end, 2 samples : total 4GB)} the running time is approximatelly 4hours. Similar with most NGS data analysis, most time consuming part within BioVLAB-MMIA-NGS workflow is currently RNAseq data process (Tophat + Cufflinks).
Trouble Shooting
- Java Runtime Environment Security issue
Recent JAVA updates (updated on 1/15/2014, version JAVA 7 Update 51) increased default security level related with Java Web Start. This cauases JNLP based applications in BioVLAB-MMIA-NGS (XBaya, Cytoscape, IGV), which run automatically downloading libraries and storing them to user system, are blocked as default. If you use the latest version of JRE and if your system encounters JAVA security issue, follow simple step by step solution described belows. Adjusting security level from 'High' to 'Medium' changes the system status from 'automatically block JNLP based applications' to 'Asking to user', so it doesn't harm your system (This was default in previous JRE version). At the same time, we are currently working with Apache Airavata group to resolve this issue. Details about recent JAVA security change, click HERE
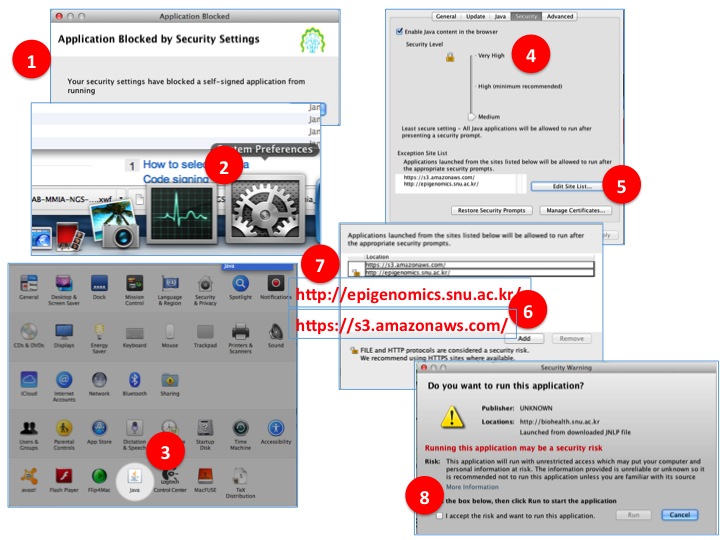
- Step by Step to resolve Java security issue for Mac OSX users.
(1) This is popup message by Java security update when the application is blocked.
(2) Open control panel.
(3) Open JAVA control panel.
(4) Click Security tab. SET SERUCITY LEVEL TO 'MIDIUM'
(5) Click 'Edit site list'.
(6) Click 'Add.
(7) Add 'http://epigenomics.snu.ac.kr/' and 'https://s3.amazonaws.com/' as in the figure
(8) Retry to lauach the BioVLAB-MMIA-NGS XBaya and now you will see warning popup instead of application blocked message. Check accept.
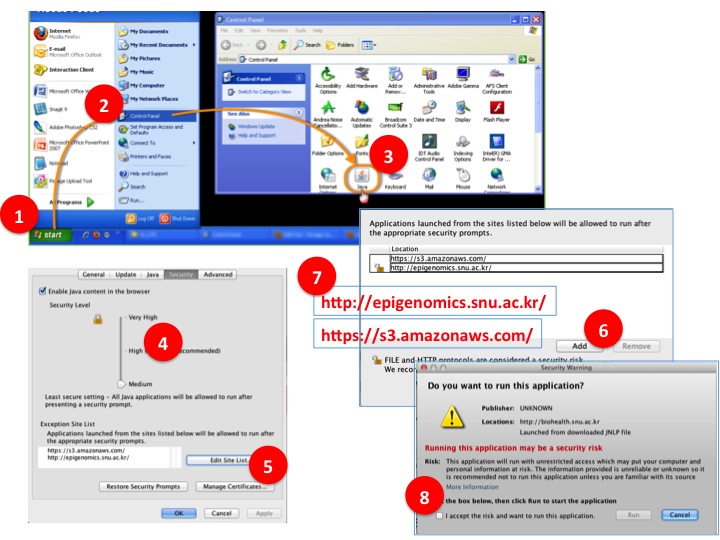
- Step by Step to resolve Java security issue for Windows users.
(1) Click 'Start' button.
(2) Open Control panel.
(3) Open JAVA control panel.
(4) Click Security tab. SET SERUCITY LEVEL TO 'MIDIUM'
(5) Click 'Edit site list'.
(6) Click 'Add.
(7) Add 'http://epigenomics.snu.ac.kr/' and 'https://s3.amazonaws.com/' as in the figure
(8) Retry to lauach the BioVLAB-MMIA-NGS XBaya and now you will see warning popup instead of application blocked message. Check accept.